Project leader:
Dr. Cátia Igreja
Department:
IV - Evolutionary Biology
Director:
Ralf. J. Sommer
Office:
Maria Gölz
Max-Planck-Ring 9
D-72076 Tübingen, Germany
Phone: +49 7071 601 441
Fax: +49 7071 601 498
Environmental changes shape developmental and behavioral traits in the nematode Pristionchus pacificus. The molecular mechanisms associated with such phenotypic plasticity are under scrutiny at the Sommerlab. I have spent the last years studying how eukaryotic cells control messenger RNA (mRNA) and protein levels. My research focused on how different protein complexes assemble and operate in the regulation of translation, and coordinate protein synthesis with mRNA stability. As a new member and group leader of the Department for Integrative Evolutionary Biology, I intent to apply my knowledge on RNA metabolism and proteins to investigate the regulation of gene expression and plastic traits in P. pacificus. Our approach is interdisciplinary, combining biochemistry, molecular and cellular biology with in vivo manipulation and characterization.
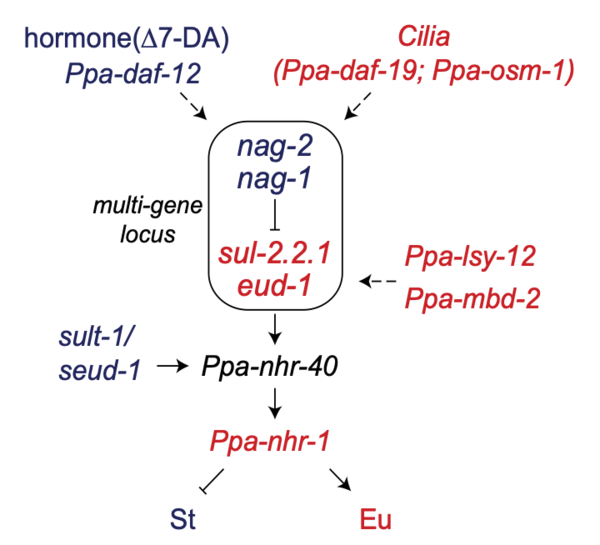
Previous work from the laboratory has identified key players in the control of mouth-form in P. pacificus. The mouth-form gene regulatory network (Fig. 1) is enriched in genes encoding proteins with distinct enzymatic activity: sulfatases (eud-1, sul-2.2.1), sulfotransferase (sult-1), N-acetylglucosaminidases (nag-1, nag-2) and histone acetyltransferase (lsy-12). To date, our knowledge on the expression, activity and substrates of these enzymes is still limited. To understand their role in the acquisition of a specific mouth-form, we intent to characterize the expression and functions of different enzymes in P. pacificus. We will apply several biochemical, genetic, molecular and proteomic approaches to answer the following questions:
In this project we will explore the time and spatial regulation of the enzymes associated with mouth-form dimorphism, uncover regulatory mechanisms controlling enzyme activity and expression in cells, and identify potential substrates.
We are actively recruiting a graduate student to work on the biochemistry and post-translational modification of enzymes. If you are interested contact us under: catia.igreja[at]tuebingen.mpg.de
Weber, R., Chung, M.Y., Keskeny, C., Zinnall, U., Landthaler, M., Valkov, E., Izaurralde, E., and Igreja, C. (2020). 4EHP and GIGYF1/2 Mediate Translation-Coupled Messenger RNA Decay. Cell Rep 33, 108262. https://doi.org/10.1016/j.celrep.2020.108262
Rasch, F., Weber, R., Izaurralde, E., and Igreja, C. (2020). 4E-T-bound mRNAs are stored in a silenced and deadenylated form. Genes Dev. http://www.genesdev.org/cgi/doi/10.1101/gad.336073.119
Peter, D., Ruscica, V., Bawankar, P., Weber, R., Helms, S., Valkov, E., Igreja, C., and Izaurralde, E. (2019). Molecular basis for GIGYF-Me31B complex assembly in 4EHP-mediated translational repression. Genes Dev 33, 1355-1360. http://www.genesdev.org/cgi/doi/10.1101/gad.329219.119
Ruscica, V., Bawankar, P., Peter, D., Helms, S., Igreja, C., and Izaurralde, E. (2019). Direct role for the Drosophila GIGYF protein in 4EHP-mediated mRNA repression. Nucleic Acids Res 47, 7035-7048. https://doi.org/10.1093/nar/gkz429
Gruner, S., Weber, R., Peter, D., Chung, M.Y., Igreja, C., Valkov, E., and Izaurralde, E. (2018). Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast. Nucleic Acids Res 46, 6893-6908. https://doi.org/10.1093/nar/gky542
Peter, D., Weber, R., Sandmeir, F., Wohlbold, L., Helms, S., Bawankar, P., Valkov, E., Igreja, C., and Izaurralde, E. (2017). GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression. Genes Dev 31, 1147-1161. http://www.genesdev.org/cgi/doi/10.1101/gad.299420.117
Gruner, S., Peter, D., Weber, R., Wohlbold, L., Chung, M.Y., Weichenrieder, O., Valkov, E., Igreja, C., and Izaurralde, E. (2016). The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol Cell 64, 467-479. https://doi.org/10.1016/j.molcel.2016.09.020
Peter, D., Weber, R., Kone, C., Chung, M.Y., Ebertsch, L., Truffault, V., Weichenrieder, O., Igreja, C., and Izaurralde, E. (2015b). Mextli proteins use both canonical bipartite and novel tripartite binding modes to form eIF4E complexes that display differential sensitivity to 4E-BP regulation. Genes Dev 29, 1835-1849. http://www.genesdev.org/cgi/doi/10.1101/gad.269068.115
Peter, D., Igreja, C., Weber, R., Wohlbold, L., Weiler, C., Ebertsch, L., Weichenrieder, O., and Izaurralde, E. (2015a). Molecular Architecture of 4E-BP Translational Inhibitors Bound to eIF4E. Mol Cell. https://doi.org/10.1016/j.molcel.2015.01.017
Igreja, C., Peter, D., Weiler, C., and Izaurralde, E. (2014). 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat Commun 5, 4790. https://doi.org/10.1038/ncomms5790
Igreja, C., and Izaurralde, E. (2011). CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev 25, 1955-1967. http://www.genesdev.org/cgi/doi/10.1101/gad.17136311